HPC Campus Clusters
This page presents an overview of the High-Performance Computing Clusters at UC Merced.
As well as how to get access, logging in, file system, resource breakdown.
Currently, UC Merced has two clusters on site. They are maintained by the CIRT team. If you have any questions, feel free to contact us here.
- Pinnacles
- MERCED
The NSF-MRI funded Pinnacles cluster located in the server facility (see Research Facility below) is available for all faculty projects at NO COST! The Pinnacles cluster runs with the Rocky (8.10) operating system, and employs the Slurm job scheduler and queueing system to manage job runs.The Pinnacles cluster is equipped with the latest generation Intel Xeon Gold 6330 CPUs and NVIDIA Tesla A100 v4 40GB HBM2 GPUs.
Facility Statement
The NSF-MRI grant number #2019144 funded Pinnacles cluster has the following compute node configurations:
- 40 regular Compute nodes with 2XIntel-28-Core Xeon Gold 6330 2.0GHz - 205W, each with 256GB RAM.
- 8 regular compute nodes with 2x Intel 32-Core Xeon Gold 6530 2.1GHz - 270W, each with 256GB RAM.
- 8 High Memory nodes, 4 nodes are equipped with 2 Intel 28-core Xeon Gold 6330 CPUs running at 2.0 GHz and 1 TB of RAM. The other 4 nodes feature 2 Intel 32-core Xeon Gold 6530 CPUs running at 2.1 GHz and 1 TB of RAM, providing additional processing power for memory-intensive workloads.
- 16 GPU nodes, 8 of the nodes are equipped with 2× NVIDIA Tesla A100 PCIe v4 GPUs (40GB HBM2), and the other 8 nodes are equipped with 2× NVIDIA L40S GPUs (48GB GDDR6).
Pinnacles also has ~92TB NFS Fast Scratch Storage space for accessing large data with low latency and 1.5PB of usable long-term storage.
Relative proximity and extent of availability: The Pinnacles cluster is managed by the Office of Information Technology at UC Merced and technical support and training opportunities are available. It is available for all faculty projects at no cost. All above nodes are interconnected via HDR InfiniBand w/ RDMA for fast (100Gbits/s) and low latency (sub ms) data transfer.
How to cite
All Pinnacles users must agree to acknowledge the Pinnacles Cluster and the supporting NSF grant (NSF MRI, # 2019144) in talks, posters, manuscripts, and other forms of dissemination relying on results obtained from time on Pinnacles. An example acknowledgement section is:
This research [Part of this research] was conducted using Pinnacles (NSF MRI, # 2019144) at the Cyberinfrastructure and Research Technologies (CIRT) at University of California, Merced.
From time to time the Committee on Research Computing (CoRC) may request a report of publications and presentations authored by Pinnacles users that have included results of calculations on Pinnacles. This information may be used by CoRC in advertising and report documents, future proposals, and/or other materials related to research computing at UC Merced.
CENVAL-ARC Node Use on Pinnacles
In addition, for those who also use cenval-arc nodes, please add the citation below to support NSF grant (#2346744). An example acknowledgement section is:
This research [Part of this research] was conducted using CENVAL-ARC compute resources on the Pinnacles cluster (NSF #2346744) at the Cyberinfrastructure and Research Technologies (CIRT) at University of California, Merced.
The MERCED (Multi-Environment Research Computer for Exploration and Discovery) Cluster is a 1,872-core, Linux-based high-performance computing system. The MERCED cluster runs with the Rocky (8.10) operating system, and employs the Slurm job scheduler and queueing system to manage job runs. MERCED operates on a Recharge model, meaning users are billed per core-hour of usage. Further details on the recharge process can be found below. To apply for a MERCED account, users must have a Chart of Account (COA) number ready.
Facility Statement
MERCED is a general-purpose computing cluster located in the server facility (see Research Facility below). The cluster consists of a login node, 65 compute nodes, and 15 high memory nodes. Total CPU-core counts is 1872.
How to cite
All MERCED users must agree to acknowledge the MERCED Cluster and the supporting UC,Merced Office of Information Technology central funded MERCED in talks, posters, manuscripts, and other forms of dissemination relying on results obtained from time on MERCED. An example acknowledgement section is:
This research [Part of this research] was conducted using MERCED cluster, which is centrally funded by the University of California, Merced, and maintained by the Cyberinfrastructure and Research Technologies (CIRT) team at UC Merced.
Recharge details
MERCED recharge calculations
Total Cost ($) = # of cores x Duration (wall clock hours) x (cost per core-hour)
- A core-hour is a single compute core used for one hour (a core-hour) and 2G of RAM.
- Cost per core-hour is $0.01
why should I be willing to invest in this?
- 🔴 The MERCED cluster offers unlimited wall clock time, allowing users to run significantly longer jobs without time constraints.
- 🟢 The MERCED queue is less crowded, allowing jobs to be picked up with shorter waiting times.
- 🟡 All recharge funds will go toward cluster maintenance, including network switch replacements and hardware upkeep. The CIRT team will not profit from these contributions.
Research Facility
The Research Computing Facility, named the Borg Cube, is a 1,448 square foot pre-manufactured, self-contained, fully serviceable data center-style building comprised of two (2) transportable modular containers, one section for electrical distribution and the other to house research computing machines. The design includes three (3) 60-ton Indirect Evaporative Cooling (IEC) mechanical units to be located outside, adjacent to the structure. The Borg Cube houses twenty (20) 19"-24" racks with 51U per rack, totaling 1020U and provides 400kW of N+1 redundant power capacity at the rack bus, providing the ability to provide individual racks with either redundant or non-redundant power supply units. The design includes two (2) 500kVA PDU's each fed from separate sources backed up from dedicated 500kVA UPS which are connected to two (2) 800A rated Starline bus to account for single or dual corded customer connections. An N+1 UPS system will consist of two (2) 500kVA UPS units, each to be equipped with batteries to support its full load for 15 minutes. The mechanical design includes three (3) 60-ton IEC units totaling 633kW of cooling available. This design accounts for an additional unit (N+1), in the case of maintenance or failure of an operating unit. The design technical requirements do not list the requirement for an N+1 mechanical system as this building can be fully supported by two (2) 60-ton units; however, due to the importance of this facility, the third unit exists for operations and maintenance purposes. This approach gives the facility engineers the ability to provide maintenance on a given unit without shutting down the facility and limiting unnecessary down time. A self-contained FM-200 style suppression system along with the required detection devices are provided for fire protection and detection systems. One (1) FM-200 tank is provided for each individual module. A VESDA system is present to detect smoke at its earliest stage and to send a signal to the clean agent suppression panel. Two (2) 1500kva substations based on an N+1 design, are provided to accept separate services. Each substation is provided with a 2000A main breaker, a 1000A breaker for the current facility, and an open space for a future circuit breaker to be used to support a potential future expansion. One of the substations is provided with a 100A breaker for the electrical equipment serving the site loads.
Cluster Hardware Configuration
- Pinnacles Hardware Overview
- Merced Hardware Overview
Compute nodes: Compute nodes are where actual jobs run. There are three types of compute nodes on Pinnacles.
- 48 Regular memory (RM) CPU nodes with 256GB RAM
- 8 Big memory CPU nodes (bigmem) with 1TB RAM
- 16 GPU Nodes
- 8 GPU nodes with NVIDIA A100 GPUs
- 8 GPU nodes with NVIDIA L40S GPUs
| CPU node | RM node | bigmem node |
|---|---|---|
| Number of nodes | 40 | 4 |
| CPU | 2 Intel 28 core Xeon Gold 6330 | 2 Intel 28 core Xeon Gold 6330 |
| RAM | 256GB | 1TB |
| Node-local storage | 1TB NVMe Data Center Solid State Drive (SSD) | 1TB NVMe Data Center Solid State Drive (SSD) |
| Network | ConnectX-6 VPI adapter card, HDR 100 InfiniBand (100Gb/s) and 100GbE, single-port QSFP56, PCIe3/4 x16 Slot | ConnectX-6 VPI adapter card, HDR 100 InfiniBand (100Gb/s) and 100GbE, single-port QSFP56, PCIe3/4 x16 Slot |
gpu GPU node | |
|---|---|
| Number | 8 |
| GPU per node | 2x NVIDIA Tesla A100 PCIe v4 40GB HBM2 Passive Single GPU |
| CPU | 2x Intel 28-Core Xeon Gold 6330 |
| RAM | 256GB |
| Node-local storage | 1TB M.2 NVMe Data Center Solid State Drive (110mm) |
| Network | ConnectX-6 VPI adapter card, HDR-100 IB (100Gb/s) and 100GbE, single-port QSFP56, PCIe3/4 x16 Slot |
cenvalarc CPU Nodes | cenvalarc.compute - CPU Node | cenvalarc.bigmem - bigmem node | OSG* |
|---|---|---|---|
| Number of nodes | 9 | 4 | 4 |
| CPU | 2x Intel 32-Core Xeon Gold 6430 2.1GHz - 270W. Or 2× Intel Xeon Gold 6530, 32-Core, 2.3 GHz, 225 W. | 2x Intel 32-Core Xeon Gold 6430 2.1GHz - 270W | 2x Intel 32-Core Xeon Gold 6430 2.1GHz - 270W. Or 2× Intel Xeon Gold 6530, 32-Core, 2.3 GHz, 225 W. |
| RAM | 256GB | 1TB | 256GB |
| Node-local storage | 1TB M.2 NVMe Data Center Solid State Drive (110mm). Or 2× 960 GB 2.5″ NVMe PCIe SSDs per node | 1TB M.2 NVMe Data Center Solid State Drive (110mm) | 1TB M.2 NVMe Data Center Solid State Drive (110mm). Or 2× 960 GB 2.5″ NVMe PCIe SSDs per node |
| Network | ConnectX-6 VPI adapter card, HDR-100 IB (100Gb/s) and 100GbE, single-port QSFP56, PCIe3/4 x16 Slot | ConnectX-6 VPI adapter card, HDR-100 IB (100Gb/s) and 100GbE, single-port QSFP56, PCIe3/4 x16 Slot | ConnectX-6 VPI adapter card, HDR-100 IB (100Gb/s) and 100GbE, single-port QSFP56, PCIe3/4 x16 Slot |
| Assigned Nodes | node[074-076],node[081-086] | hmnode[007-010] | node[077-080] |
* OSG Nodes are only accessable via test partition. To learn more information about the Open Science Grid (OSG) can be found here.
cenvalarc.gpu GPU node | L40S Nodes | H200 w/ NVLINK Nodes |
|---|---|---|
| Number of nodes | 8 | 4 |
| GPU per node | 2× NVIDIA L40S (48GB GDDR6) | 2× NVIDIA H200 NVL (141GB HBM3) |
| SLURM GRES | gpu:l40s:2 | gpu:nvidia_h200_nvl:2 |
| CPU | 2× Intel 32-Core Xeon Gold 6530 2.1GHz - 270W | 2× Intel 32-Core Xeon Gold 6530 2.1GHz - 270W |
| RAM | 256GB | 256GB |
| Node-local storage | 1TB M.2 NVMe Data Center SSD (110mm) | 1TB M.2 NVMe Data Center SSD (110mm) |
| Network | ConnectX-6 VPI, HDR-100 IB (100Gb/s), single-port QSFP56, PCIe3/4 x16 | ConnectX-6 VPI, HDR-100 IB (100Gb/s), single-port QSFP56, PCIe3/4 x16 |
| constraint | --gres=gpu:l40s:<number of gpu> | --gres=gpu:nvidia_h200_nvl:<number of gpus> |
| Assigned Nodes | gnode[017-024] | gnode[026-029] |
MERCED hosts 66 CPU compute nodes including 25 high memory nodes. Please be aware that the nodes among MERCED cluster are multi-generational, meaning that the CPU processors from different nodes are having different features, the table shows below listed detailed node information. Users may experience relative big performance variations when running the same jobs on different nodes.
The table below listed all MERCED cluster CPU compute nodes features, and their processors generations.
| Nodes | feature | RAM | Total cores per nodes | InfiniBand (IB) |
|---|---|---|---|---|
| 33-43 | Broadwell,avx2,E5-2650_v4,local scratch 932GB | 128GB | 24 | yes |
| 44 | Broadwell,avx2,E5-2650_v4,local scratch 932GB | 112GB | 24 | yes |
| 45-60 | Broadwell,avx2,E5-2650_v4,local scratch 932GB | 257GB | 24 | yes |
| 61-72 | Broadwell,avx2,E5-2650_v4,local scratch 447GB | 257GB | 24 | yes |
| 73-76, 79-88 | Broadwell,avx2,E5-2650_v4,local scratch 932GB | 128GB | 24 | yes |
| 77 | Broadwell,avx2,E5-2650_v4,no local scratch | 128GB | 24 | yes |
| 89-104 | Skylake,sse4.2,avx,avx2,avx512,Gold_6130, no local scratch | 191GB | 32 | yes |
| 105-114 | cascadelake,sse4.2,avx,avx2,avx512,Gold_6230, no local scratch | 191GB | 40 | yes |
How to Request an Account
- Pinnacles Account Process
- MERCED Account Process
UC Merced Faculty Principal Investigators (PIs) can request access to Pinnacles cluster. All student user accounts on Pinnacles cluster must associate with UC Merced PIs.
UC Merced Principal Investigators (PIs) or other researchers request Pinnacles account here.
Click Here to View a Visual Guide for Creating an Account for Pinnacles
Requesting Access to Pinnacles Process.
- UC Merced Principal Investigators (PIs) or other researchers request Pinnacles account here.
- For new account group project applications, PIs please also make sure to complete the export control form, if the PI has not done one before.
- Once the form is completed, please attach the form to the request ticket scene in the following steps.
- Click
Request Service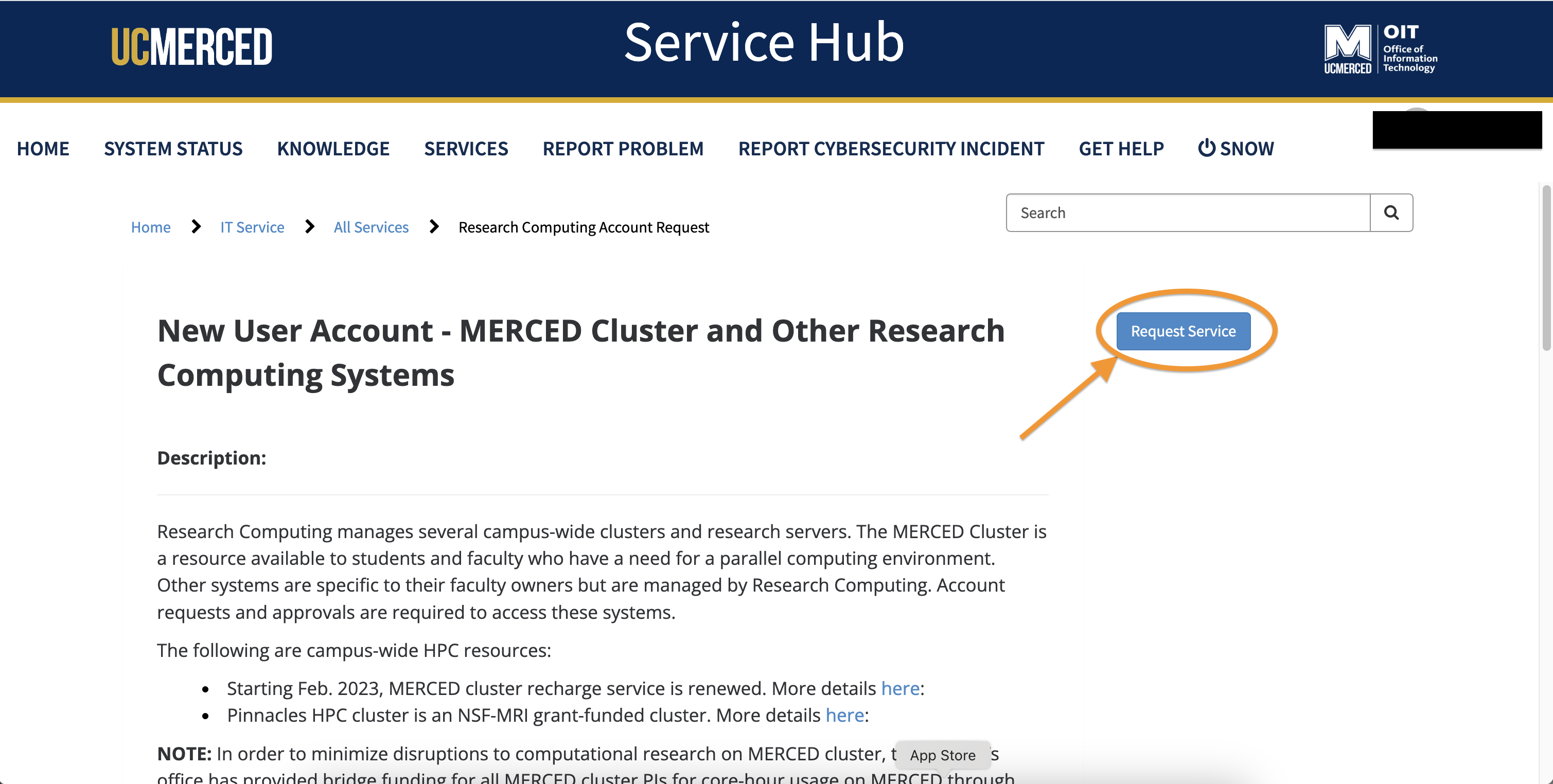
- Begin to populate all the required information.
- At the question regarding PI Status. Typically only Professors are PIs, their students and post-docs would select
Noat this question.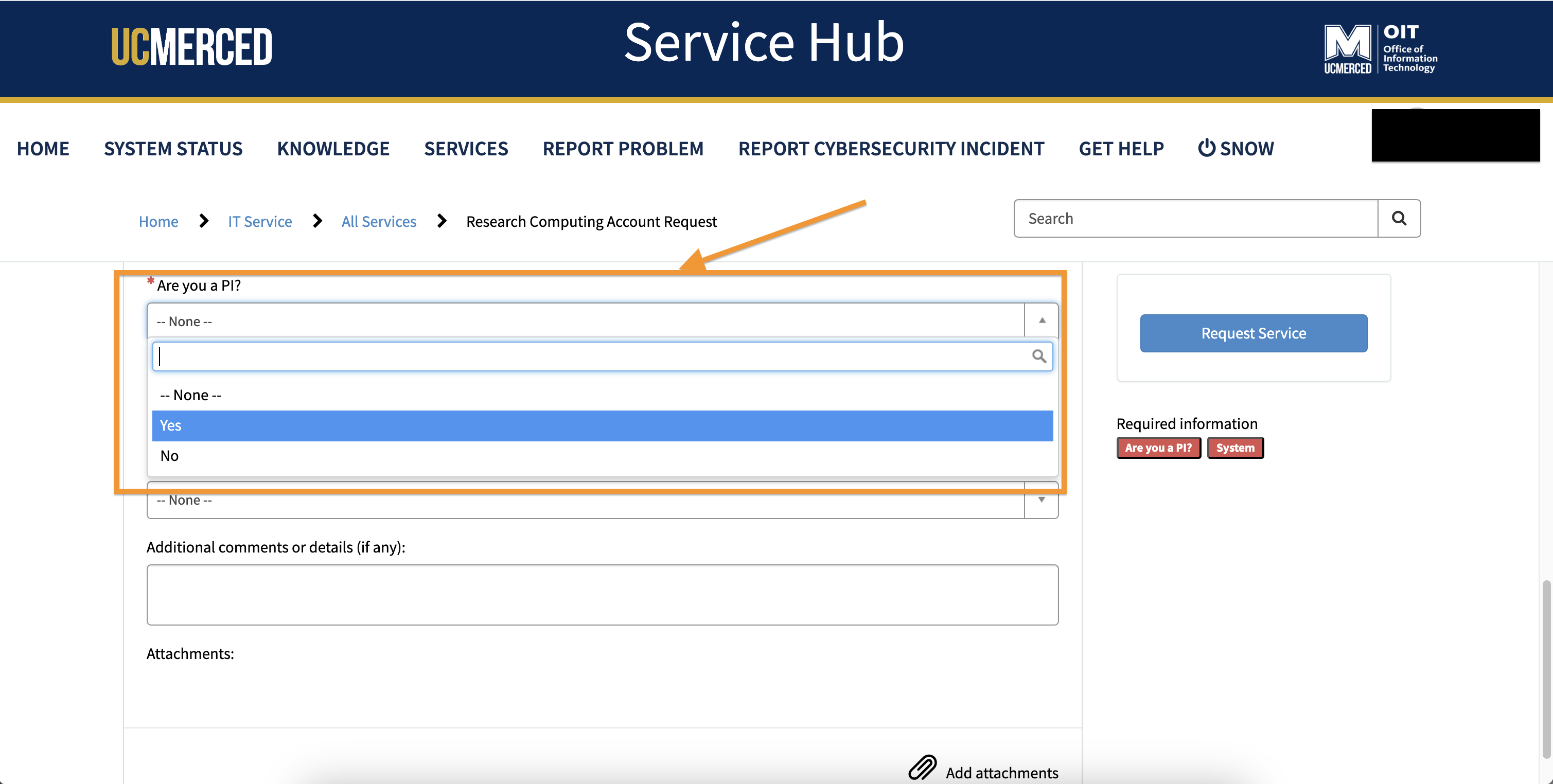
- At the question regarding PI Status. Typically only Professors are PIs, their students and post-docs would select
- For selecting the system, from the drop-down, click
pinnacles.ucmerced.edu (Free Cluster)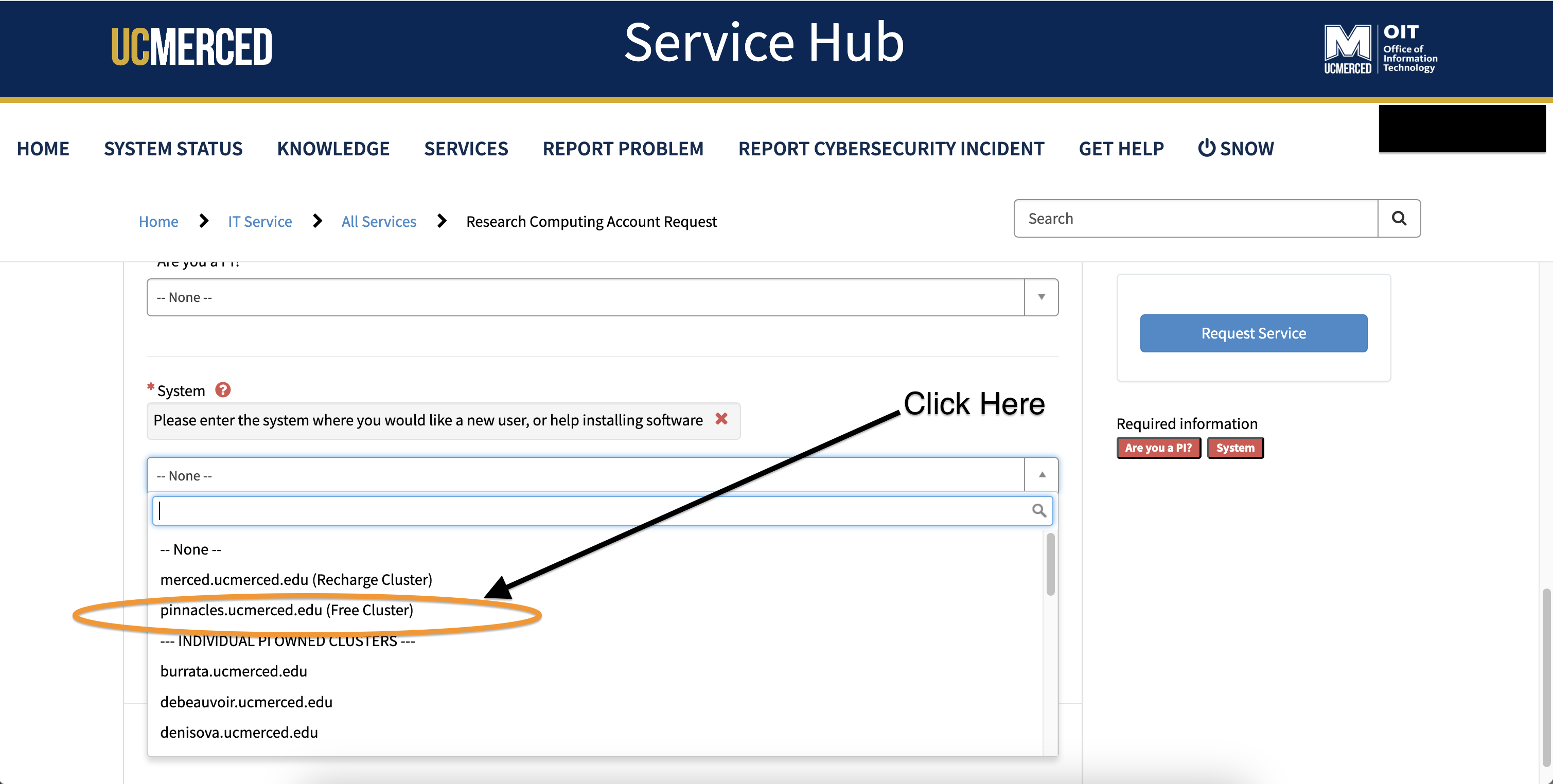
- Add any other additonal comments or information, you believe will be helpful for the requesting an account process.
- Click
Request Service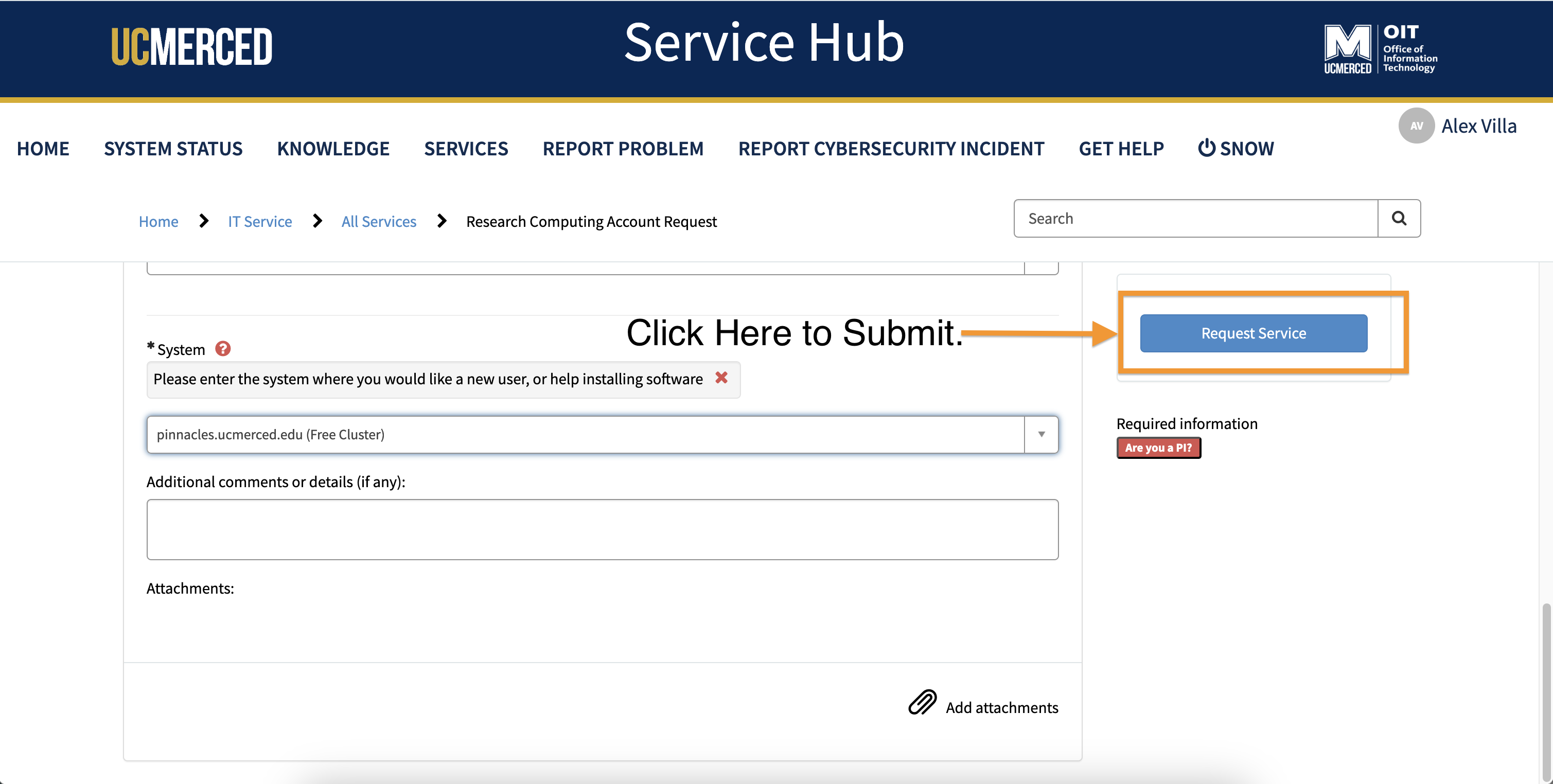
UC Merced Faculty Principal Investigators (PIs) can request access to MERCED cluster. All student user accounts on MERCED cluster must associate with UC Merced PIs. UC Merced Principal Investigators (PIs) or other researchers request MERCED account here.
MERCED is a recharge cluster, and will require all accounts to have an associated COA Number submitted at time of account request.
Click Here to View a Visual Guide for Creating an Account for MERCED
Requesting Access to MERCED Process.
- UC Merced Principal Investigators (PIs) or other researchers request MERCED account here.
- For new account group project applications, PIs please also make sure to complete the export control form, if the PI has not done one before.
- Once the form is completed, please attach the form to the request ticket scene in the following steps.
- Click
Request Service
- Begin to populate all the required information.
- At the question regarding PI Status. Typically only Professors are PIs, their students and post-docs would select
Noat this question.
- At the question regarding PI Status. Typically only Professors are PIs, their students and post-docs would select
- For selecting the system, from the drop-down, click
merced.ucmerced.edu (Recharge Cluster)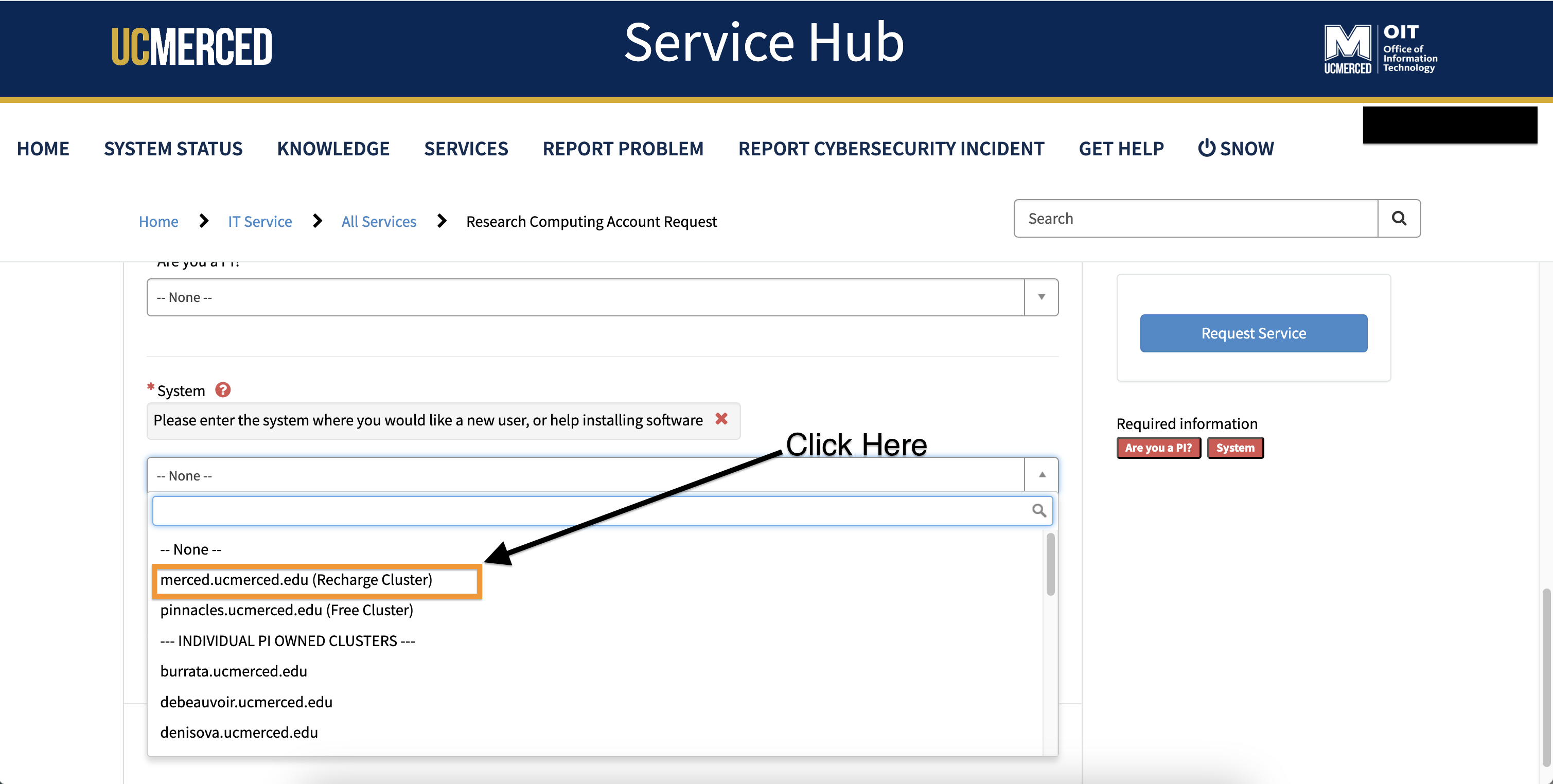
- Because MERCED is a Recharge Cluster, please attach a valid COA number. Without a COA number, the account request will be denied.
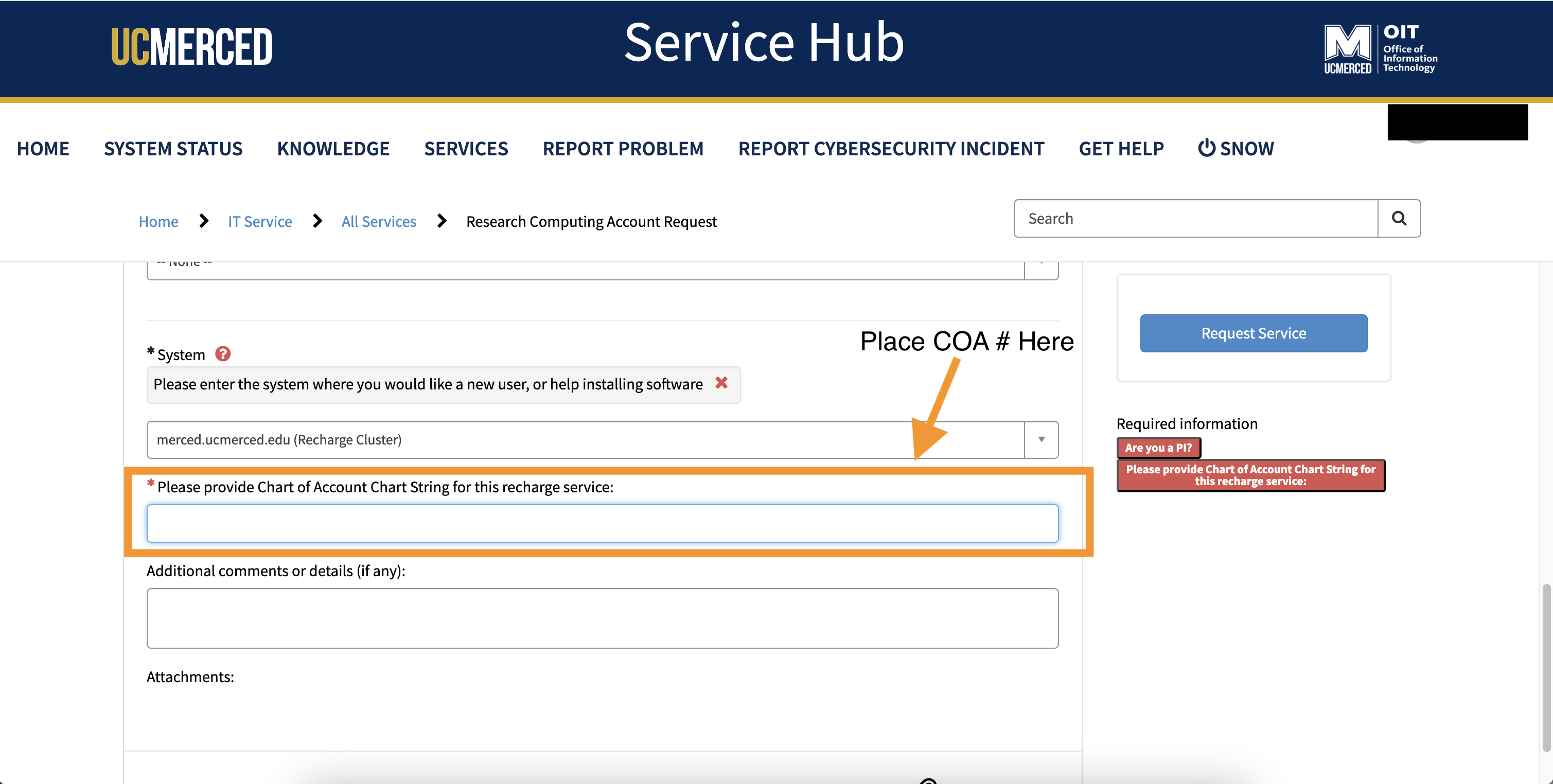
- Add any other additional comments or information, you believe will be helpful for the requesting an account process.
- Click
Request Service
Centralized login
Open OnDemand Login
For users seeking to access access Pinnacles and MERCED cluster via the web-based GUI, Open OnDemand. Please refer to this page here for accessing and making the most of the Open OnDemand Interface.
If connecting via SSH to the clusters from on campus, connect to eduroam or UCM CatNet. Otherwise ensure you are connected to the Campus VPN.
For users who want a traditional interface experience and ssh experience that can still be done as before, using the below method.
Login nodes
The standard method for connecting to a remote machine is through Secure Shell (ssh) commands. Pinnacles and MERCED are accessed via a centralized login. This means that once a user logs into one of the login nodes, they will be able to access both the MERCED and Pinnacles clusters. Users applying for a Pinnacles account can begin the application process here, and Pinnacles is FREE to use within the campus. However, to access the MERCED cluster, users must provide a COA account number and enter the number during the MERCED account application process.
Currently, we have three login nodes, and users can expect to be connected to either rclogin01, rclogin02, or rclogin03. Do not run computationally intensive processes on the login nodes. These nodes are appropriate for tasks such as file preparation/editing, compiling, simple analyses, and other low-computation activities. For more resource-intensive work, submit jobs to the cluster using the available queue system. Additionally, users can connect to a remote machine using an X-terminal (XQuarz or X11) forwarding (see example command below) to run graphics-based programs like gnuplot, gimp, etc.
Connect to the clusters
On Mac and Linux you can use the built-in terminal application; on Windows you can use MobaXterm to open a terminal, and type the following command, but replace <username> to your UCMID.
ssh <username>@login.rc.ucmerced.edu
X11 forwarding
Mac OS:
Prerequisite: Install XQuartz
Then open XQuartz through open -a XQuartz in the terminal or other CLI. Then type the following command.
ssh -X <username>@login.rc.ucmerced.edu
-X: Enables X11 forwarding.
Windows users
MobaXterm includes an integrated X11 server, so no additional installation of X11 software is needed.
- Start a New SSH Session with X11 Forwarding
- In the top-left corner, click on the "Session" button.
- Choose "SSH" from the available options
- Configure the SSH Session
- In the Remote Host field, enter the address of the remote server (e.g., remote.server.com)
- Ensure the "Specify username" box is checked, then enter your username for the remote server
- Check the box that says "X11-forwarding". This option enables X11 forwarding for your session
File systems and storage
There are 2 folders (data and scratch) located in HOME that users will start with.
MERCED and Pinnacles have now been merged into a centralized system, allowing them to share the same file systems. We have also increased the quota for the data, scratch, and HOME directories. Please note that there is a 7-day grace period once the soft quota limit is reached.
| Folder | soft quota | hard quota |
|---|---|---|
HOME | 70G | 75G |
data | 500G | 512G |
scratch | 500G | 512G |
The scratch folder is purged periodically when the overall system storage reaches 85% of capacity or higher. Please back-up your data to somewhere safe frequently.
Queue Information
- Pinnacles Queue Information
- MERCED Queue Information
Pinnacles Cluster is the default cluster that is free and accessible to all users and has 6 public queues.
| Public Queues(Available to all users) | Max Wall Time | Default Time | Max Nodes per Job | Max # of jobs that can be submitted |
|---|---|---|---|---|
| ^test | 1 hour | 5 min. | 2 nodes | 1 |
| bigmem | 3 days | 1 hrs | 2 nodes | 2 |
| gpu | 3 days | 1 hrs | 2 nodes | 4 |
| *short | 6 hours | 1 hrs | 4 nodes | 12 |
| medium | 1 day | 6 hrs | 4 nodes | 6 |
| long | 3 days | 1 day | 4 nodes | 3 |
| cenvalarc.compute | 3 day | 1 day | 4 nodes | 3 |
| cenvalarc.bigmem | 3 day | 1 day | 2 nodes | 2 |
| cenvalarc.gpu | 3 day | 1 day | 2 nodes | 4 |
short queue is the default queue for all jobs submitted without specifying which queue job must run on
^test queue has access to all node types use constraints to test on specific types. Ex:
#SBATCH --constraint=gpu,bigmem
Access to GPUs also requires
#SBATCH --gres=gpu:X
MERCED is the Recharge Cluster.
| Public Queues(Available to all users) | Max Wall Time | Default Time | Max Nodes per Job | Max # of jobs that can be submitted |
|---|---|---|---|---|
| bigmem | 5 days | 1 hr | 2 nodes | 6 |
| test^ | 1 hour | 5 min. | 2 nodes | 1 |
| *compute | 5 days | 1 hr | 2 nodes | 6 |
#SBATCH -M merced must always be used to submit a job to MERCED cluster
^ test queue has access to all node types use constraints to test on specific types.
compute queue is the default queue for all jobs submitted
Global Modules on Pinnacles and MERCED
Pinnacles and MERCED already come with a collection of global modules or softwares that do not need to be individually installed by the user. The module system allows for the loading and unloading of a specific module. Users will make use of avail, load, list, unload, and swap. A table describing each of these Modules options is given below.
A complete guide to using modules can be found via man module.
| Command | Description |
|---|---|
module avail | This command lists all available modules |
module load <mod_name> | This command loads the environment corresponding to <mod_name> |
module list | This command provides a list of all modules currently loaded into the user environment |
module unload <mod_name> | This command unloads the environment corresponding to <mod_name> |
module swap <mod_1> <mod_2> | This command unloads the environment corresponding to <mod_1> and loads to <mod_2> |
- Pinnacles and MERCED Global Modules
- MERCED Only Modules
Click Here to Expand to View the List.
admin/0.0.1 gaussian/gdv-20170407-i10+ mpfr/4.2.0 r-biobase/2.50.0
amber/20-devel gaussian/gdv-20210302-j15 (D) mpich/3.4.2-gcc-8.4.1 r-ctc/1.64.0
amber/20 (D) gcc/8.5.0 mpich/3.4.2-intel-2021.4.0 r-deseq2/1.30.0
anaconda3/2021.05 gcc/11.2.0 mpich/3.4.2-nvidiahpc-21.9-0 (D) r-edger/3.32.1
anaconda3/2023.09-0 (D) gcc/12.2.0 (D) multiqc/1.7 r-fastcluster/1.1.25
angsd/0.940 git/2.37.0 multiwfn/3.8 r-glimma/2.0.0
apbs/3.4.1 glpk/4.65 mvapich2/2.3.6-gcc-8.4.1 r-goplot/1.0.2
awscli/1.16.308 gmp/6.2.1 mvapich2/2.3.6-intel-2021.4.0 (D) r-goseq/1.42.0
bamtools/2.5.1 gnuplot/5.4.2 ncbi-blast+/2.12.0 r-gplots/3.1.1
bamutil/1.0.15 grace/5.1.25 netlib-lapack/3.9.1 r-qvalue/2.22.0
bbmap/39.06 gromacs/2021.3 netlib-xblas/1.0.248 r-rots/1.18.0
bcftools/1.12 gromacs/2022.3 nvidiahpc/21.9-0 r-sm/2.2-5.6
bcftools/1.14 (D) gromacs/2023.1 (D) octopus/13.0 r-tidyverse/1.3.0
bcl2fastq2/2.20.0.422 gsl/2.7 octopus/14.1 (D) r/4.1.1
beast/1.10.4 gurobi/9.5.0 onnx/1.10.1 r/4.2.2 (D)
beast2/2.6.4 hdf5/1.10.7-intel-2021.4.0 openbabel/3.0.0 raxml-ng/1.2.0
bedtools2/2.30.0 hdf5/1.14.1-2 (D) openblas/0.3.18 rclone/1.59.1
berkeleygw/3.0.1-intel-mvapich2 ibamr/0.8.0-testing openblas/0.3.21 (D) repeatmodeler/1.0.11
berkeleygw/3.0.1-intel-2021.4.0 ibamr/0.12.0-debug opencarp/8.1 rsem/1.3.1
berkeleygw/3.0.1 ibamr/0.12.0-opt openjdk/1.8.0_265-b01 salmon/1.4.0
berkeleygw/4.0-mvapich2-oneapi (D) ibamr/0.13.0-debug openjdk/11.0.20 samtools/1.13
blast-plus/2.12.0 ibamr/0.13.0-opt (D) openjdk/17.0.5_8 (D) scalapack/2.1.0
bowtie/1.3.0 intel/oneapi openmpi/3.1.3-gcc schrodinger/2022-1
bowtie2/2.4.2 interproscan/5.55-88.0 openmpi/3.1.6-gcc-8.4.1 schrodinger/2022-3 (D)
braker/2.1.6 ior/3.3.0 openmpi/3.1.6-intel-2021.4.0 sickle/1.33
butterflypack/2.0.0 iq-tree/2.1.3 openmpi/3.1.6-nvidiahpc-21.9-0 singularity/3.8.3
bwa-mem2/2.2.1 jellyfish/2.2.7 openmpi/4.0-merced-test smalt/0.7.6
bwa/0.7.17 julia/1.7.3 openmpi/4.0.6-gcc-8.4.1 sombrero/2021-08-16
casacore/3.4.0 julia/10.1.1 (D) openmpi/4.0.6-intel-2021.4.0 spiral/8.2.0
cgal/5.0.3 kallisto/0.46.2 openmpi/4.0.6-nvidiahpc-21.9-0 srilm/1.7.3
cmake/3.21.4 lammps/20210310+kokkos+cuda openmpi/4.1.1-gcc-8.4.1 stacks/2.53
cmaq/5.3.1 lammps/20210310+user-omp+kokkos openmpi/4.1.1-intel-2021.4.0 star/2.7.11b
collier/1.2.5 lammps/20210310 openmpi/4.1.4-gcc-12.2.0+cuda stata/17
cuda/10.2.89 lammps/20220107+ml-quip openmpi/4.1.4-gcc-12.2.0 (D) stata/18 (D)
cuda/11.0.3 lammps/20220107 orca/5.0.1 stringtie/2.2.3
cuda/11.4.0 lammps/20230208 (D) orthofinder/2.5.2 subversion/1.14.1
cuda/11.5.0 latte/1.2.2 perl-db-file/1.840 suite-sparse/5.13.0
cuda/11.8.0 lftp/4.9.2 perl-uri/1.72 tcl/8.5.19-gcc-8.5.0
cuda/12.3.0 (D) libraries perl/5.34.0 terachem/1.95
dakota/6.12 libtirpc/1.1.4 phyluce/1.6.7 tk/8.5.19-gcc-8.5.0
dalton/2020.0 libxc/5.2.3-gcc-12.2.0 picard/2.26.2 toolchain/scientificstack-11.2.0
elpa/2021.11.001 likwid/5.2.2+cuda pigz/2.7 transdecoder/5.5.0
emacs/27.2 likwid/5.2.2 (D) plink/1.90-beta-7.1 trimmomatic/0.39
express/1.5.2 localcolabfold/1.5.1 protobuf/3.18.0 trinity/2.12.0
fastqc/0.11.9 mathematica/12.3.1 py-numpy/1.21.4 trinity/2.15.1 (D)
ffmpeg/4.3.2 mathematica/14.0.0 (D) python/3.8.12 user-modules
fftw/3.3.10-gcc-8.5.0 matlab/r2021b python/3.11.0 (D) vcftools/0.1.14
fftw/3.3.10-intel-2021.4.0 (D) matlab/r2023a quantum-espresso/6.7-intel-test vmd/1.9.1
gate/9.0 matlab/r2024a (D) quantum-espresso/7.1 vmd/1.9.3 (D)
gatk/4.2.6 metis/5.1.0 quantum-espresso/7.2-gcc-openblas (D) wannier90/3.1.0
gaussian/g09-d01 minimap2/2.14 r-ape/5.4-1 xcrysden/1.6.2
gaussian/g16-b01 molden/6.7 r-argparse/2.0.3
Where:
D: Default Module
berkeleygw/3.0.1-intel-mvapich2 berkeleygw/4.0-mvapich2-oneapi (D)
bwa-mem2/2.2.1 openmpi/4.0-merced-test
user-modules
Where:
D: Default Module
Checking disk quota and usage
To look at your current usage amounts of HOME, data or scratch use the following command
quota -vs
This will output, in sections, the filesystem, current space usage, quota, hard limit, and other relevant information in a more readable format.
To convert the outputted megabytes to gigabytes = space(MB) divided by 1024
Checking the size of directories and content
To check the size of the current directory or any directories in it use the du command.
du command alone will output all directories, hidden as well, in real time so it will take a few moments to finish. It is recommended to execute the command with some of the following options to make the process more clear and concise.
| Option | Description |
|---|---|
-h | Displays storage values in human-readable format (e.g., KB, MB, GB) |
-s | Summarizes the size of the whole current directory |
-sh | Shows the size of the specified sub-directory |
--max-depth=N | Shows the sub-directories up to depth N, where N is a number representing the max depth |
--all | Writes counts for all files, not just directories |
--help | Displays all other options for the du command |
Example usage of du command:
- example
- output
du -h -s <directory name>
35G <directory name>
Users who submit jobs to MERCED and use the unified storage are expecting slower network communications.
- Home - Shared over 10G network from Pinnacles to Merced, connected over IB on pinnacles.
- Data - Shared over 10G network from Pinnacles to Merced, connected over IB on pinnacles.
- Scratch - Shared over 10G network from Pinnacles to Merced, connected over IB on pinnacles
Please avoid writing files directly to /tmp on the head node, as this can fill up disk space and cause issues for all users. Instead, use your personal scratch directory for temporary files. Some programs may default to using /tmp, so ensure that the appropriate scratch directory is properly configured for your code.
Disclaimer: Users are responsible for backing up all data stored on the clusters and are fully accountable for its availability. CIRT is not liable for any data loss in the event of accidents